Description
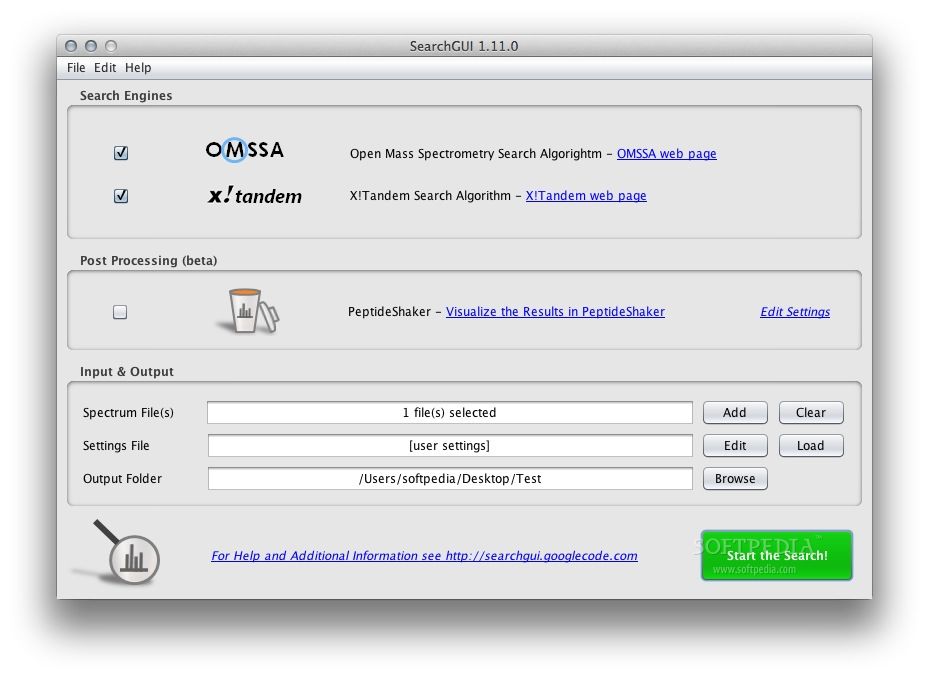
SearchGUI is an open-source project serving as a common interface for proteomics search and de novo engines, allowing you to configure and run multiple engines at the same time. It supports X! Tandem, MS Amanda, MyriMatch, MS-GF+, OMSSA, Tide, Comet, Andromeda, Novor, MetaMorpheus, and DirecTag.
Intended to simplify the task of using several search engines at the same time via a graphical user interface, SearchGUI can also be used from the command-line, and it can be added to various analysis pipelines. The program is Java-based, and it is available for Mac, Windows, and Linux.
As far as input formats are concerned, the software supports mzML and mgf files as data sources. ThermoRawFileParser is built-in, which allows you to easily convert Thermo raw files into mzML or mgf.
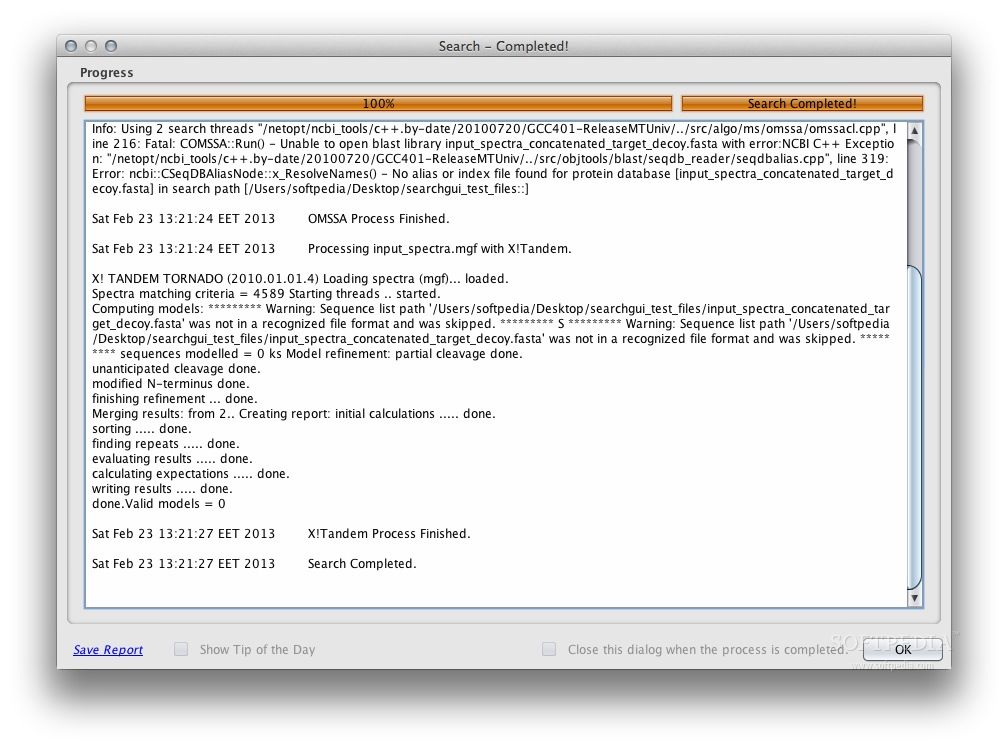
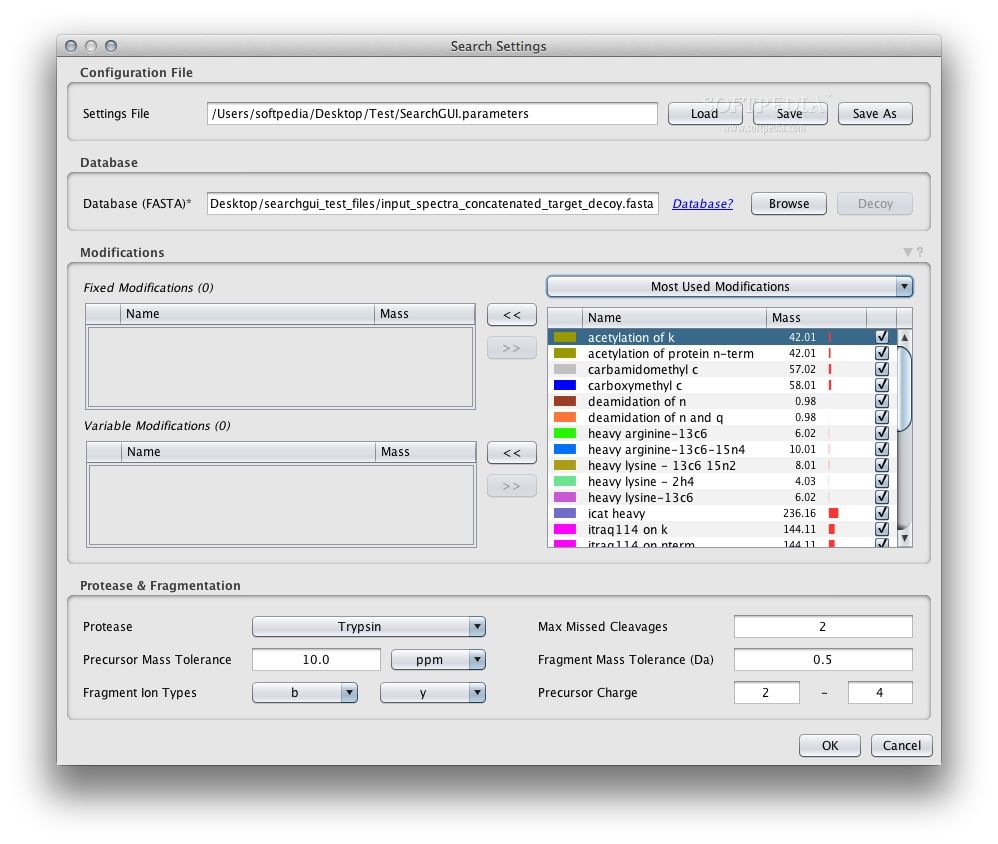
From the main user interface, you can load spectrum files and search settings, then set the preferred output folder. Pre-processing and post-processing can optionally be run. Finally, you can select the search engines you would like to use and configure them independently. Note that some engines are only available on certain operating systems.
Once the results have been provided, they can be viewed and interpreted using a specialized application, such as PeptideShaker.
User Reviews for SearchGUI FOR MAC 1
-
for SearchGUI FOR MAC
SearchGUI FOR MAC provides a seamless experience for running multiple proteomics search engines. The graphical interface simplifies configuration and execution.