Description
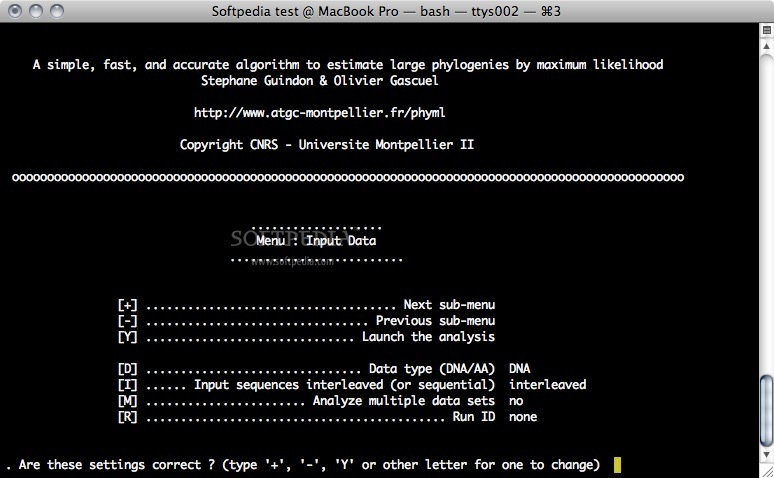
PhyML is a command line based utility that estimates maximum likelihood phylogenies from alignments of amino acid or nucleotide sequences. PhyML provides a wide range of options that were designed to facilitate standard phylogenetic analyses.
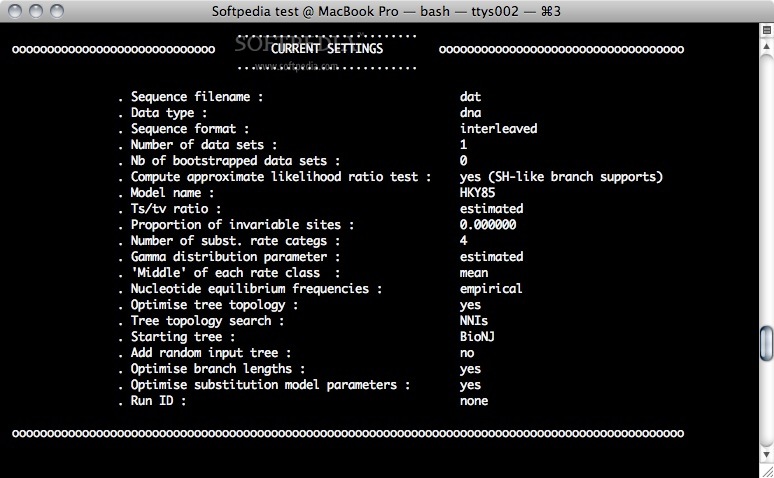
The main strengths of PhyML lies in the large number of substitution models coupled to various options to search the space of phylogenetic tree topologies, going from very fast and efficient methods to slower but generally more accurate approaches.
PhyML also implements two methods to evaluate branch supports in a sound statistical framework (the non-parametric bootstrap and the approximate likelihood ratio test).
PhyML was designed to process moderate to large data sets. In theory, alignments with up to 4,000 sequences 2,000,000 character-long can analyzed.
In practice however, the amount of memory required to process a data set is proportional of the product of the number of sequences by their length. Hence, a large number of sequences can only be processed provided that they are short.
Also, PhyML can handle long sequences provided that they are not numerous. With most standard personal computers, the “comfort zone” for PhyML generally lies around 100-200 sequences less than 2,000 character long.
User Reviews for PhyML FOR MAC 1
-
for PhyML FOR MAC
PhyML FOR MAC provides a wide range of options for phylogenetic analyses. Strengths include various substitution models and efficient tree topology search.