Description
Pathomx
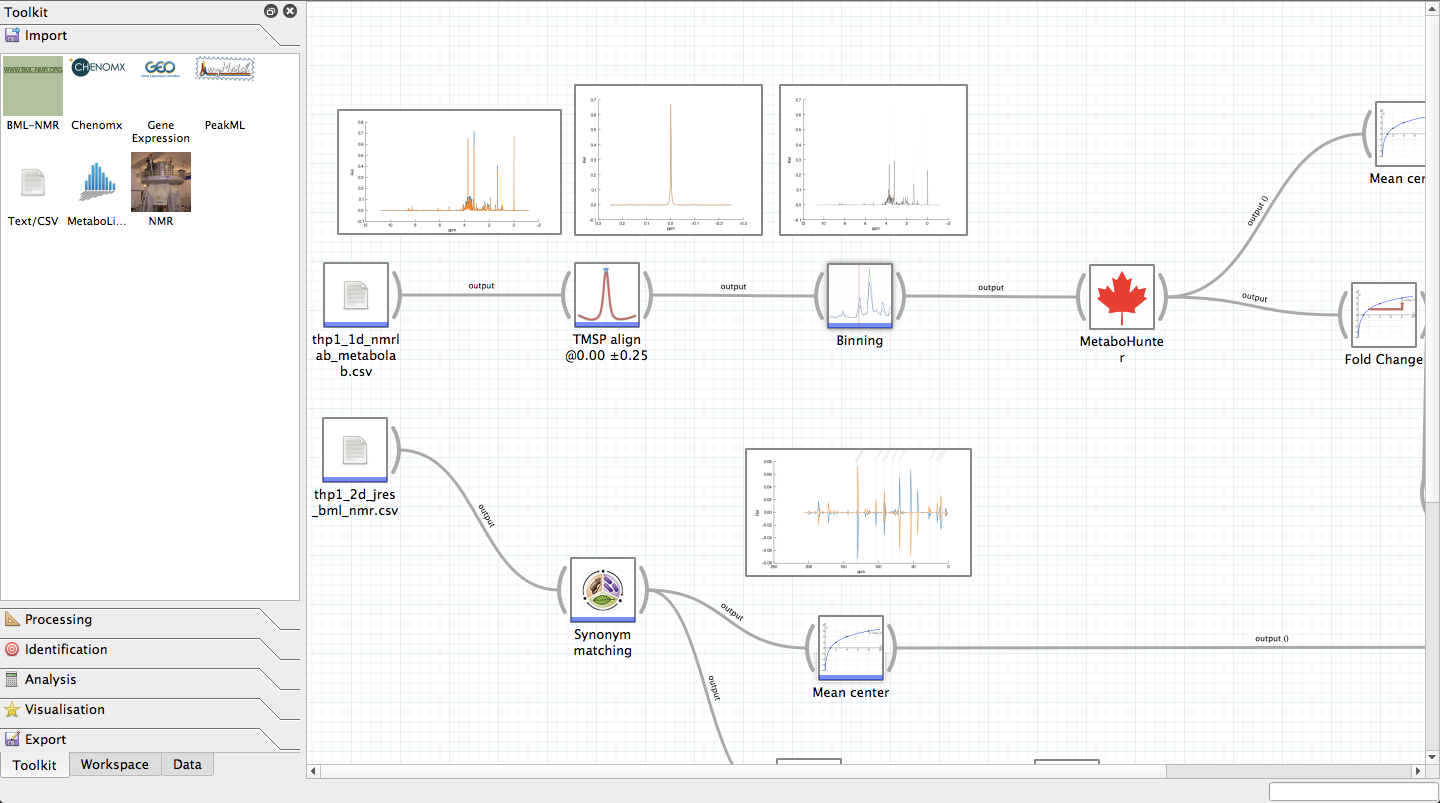
Pathomx (formerly known as MetaPath) is a cool open-source software that runs on IPython, Qt, and PyQt. It's built to be an interactive tool for looking at and analyzing metabolic data, especially for those using GNU/Linux and other POSIX operating systems. Originally, it was created to handle metabolomics data.
Key Features of Pathomx
This software has some great features! You can import and export NMR (Nuclear Magnetic Resonance) data, process spectra with options like normalization, binning, and alignment. Plus, it allows you to visualize and mine metabolic pathways! You can also bring in genomic and metabolomics data through different methods to see how they fit into specific pathways.
MetaCyc Database Integration
A big advantage of Pathomx is its connection to the MetaCyc database. This means you get handy annotations like database unification links, synonyms, related pathways, and reactions. Just remember that the MetaCyc database comes from a public API!
Powerful Tools Under the Hood
This application uses the robust MetaCyc database to explore complex datasets quickly. It has a bunch of plugins that you can customize too! For number crunching, it relies on libraries like Numpy and Scipy. For graphing, it uses Matplotlib along with nmrglue for importing NMR data. Want interactive graphs? That’s where d3.js comes in handy! And if you're into statistical analysis methods, scikit-learn has got your back.
Compatibility Across Platforms
If we peek under the hood of Pathomx, we see it's built on an IPython command shell. It works smoothly on Microsoft Windows and Mac OS X too! You can grab pre-built binary packages for those systems. For GNU/Linux users, though, you'll find it distributed as a gzipped source archive compatible with both 32-bit and 64-bit hardware platforms.
User Tips for Pathomx
A quick heads up when using Pathomx: It pulls in data from the MetaCyc pathway database as part of the BioCyc family. The MetaCyc API helps create this local database you'll be working with.
If you're ready to dive in and try out Pathomx yourself, check out this link here!
Tags:
User Reviews for Pathomx FOR LINUX 7
-
for Pathomx FOR LINUX
Pathomx FOR LINUX is a powerful tool for metabolic data analysis on GNU/Linux. Its features, plugins, and databases make it indispensable.
-
for Pathomx FOR LINUX
Pathomx is an incredible tool for metabolomics data analysis! The interface is user-friendly and the features are robust.
-
for Pathomx FOR LINUX
I love how Pathomx allows me to visualize complex metabolic pathways easily. The integration with MetaCyc is a game changer!
-
for Pathomx FOR LINUX
This app has transformed my approach to analyzing NMR data. The processing capabilities are top-notch and intuitive.
-
for Pathomx FOR LINUX
Pathomx is a must-have for anyone working with metabolic data. It's powerful, open-source, and very flexible!
-
for Pathomx FOR LINUX
The extensive support for various data formats in Pathomx makes it so versatile. Highly recommend for researchers!
-
for Pathomx FOR LINUX
Fantastic application! The ability to import genomic data alongside metabolomics is invaluable for my research.