Description
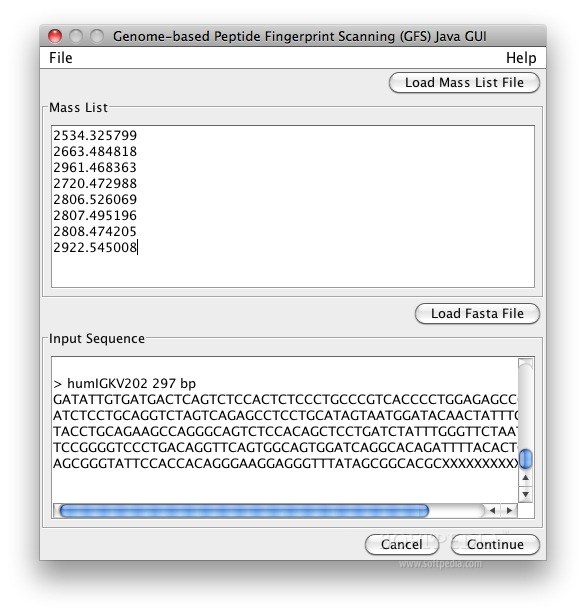
Genome-based fingerprint scanning (GFS) is a small, simple, easy to application that maps peptide mass fingerprint data directly to raw genomic sequence, enabling rapid, low-cost identification of proteins in genomes for which annotation is lacking.
An experimentally obtained peptide mass fingerprint is entered into the program, which then scans a genome sequence of interest, and outputs the most likely regions of the genome from which the mass fingerprint is derived.
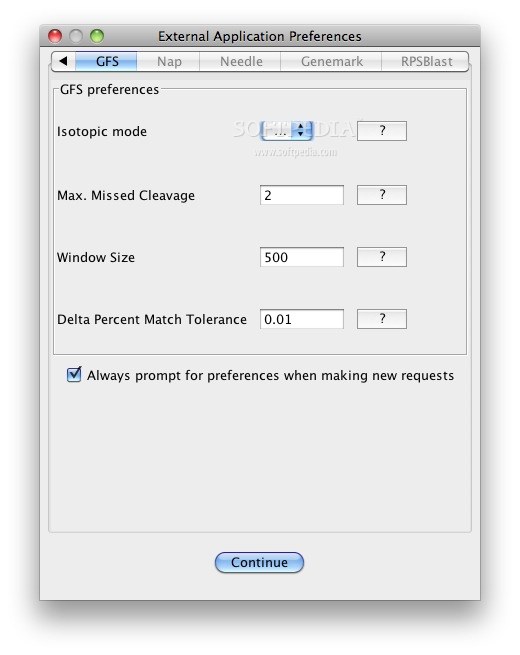
GFS first generates a theoretical mass list by translating the genome of interest in 6 reading frames (3 each on the forward and reverse strands) and digesting the resulting proteins in silico according to cleavage rules associated with the specified protease (trypsin in this case).
The algorithm then finds matches (within a given mass tolerance) between these theoretical masses and the input experimental masses.
These matches (or hits) are grouped into high-density regions on the genome that can be scored according to a number of criteria and ranked by statistical significance.
These regions are derived by scanning across the genome with a fixed-sized window and then each window is scored according to criteria detailed in the original GFS publication.
User Reviews for GFS FOR MAC 1
-
for GFS FOR MAC
GFS FOR MAC is a powerful tool for protein identification from genomes with minimal annotation. It's user-friendly and efficient.