Description
CodonW
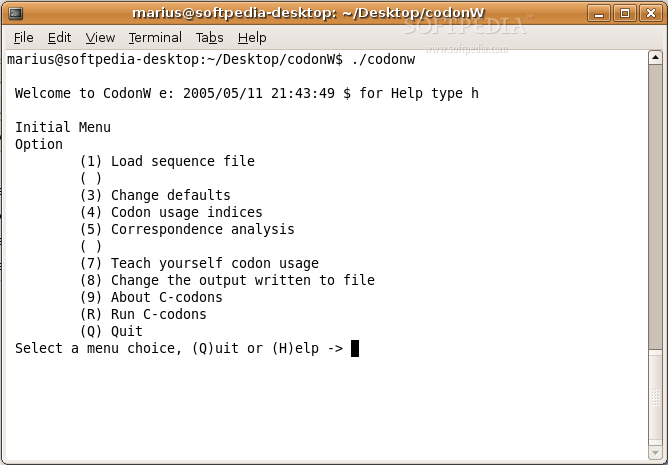
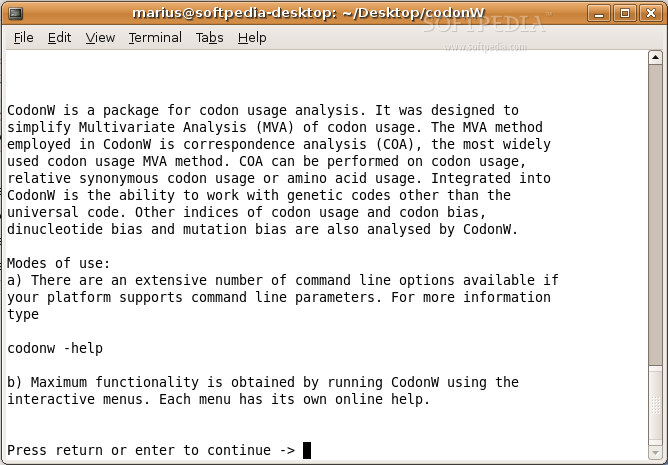
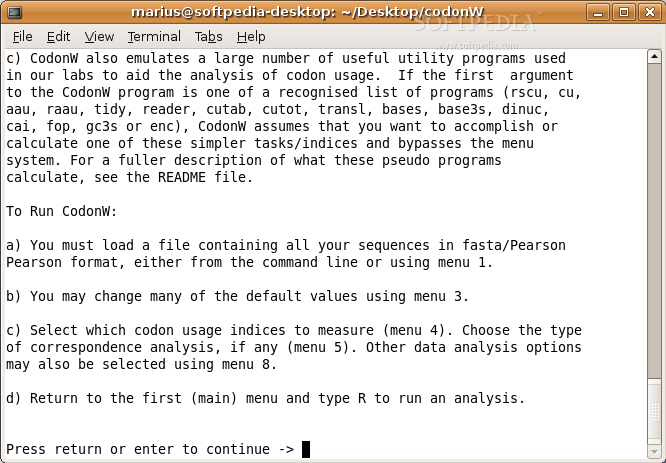
CodonW is a handy program that makes it easier to do multivariate analysis, especially when looking at codon and amino acid usage. It's also great for calculating standard indices of codon usage. Plus, it comes with both menu and command-line options, so you can pick what works best for you!
About the CodonW Project
The CodonW project was created by John Peden while he was working in Paul Sharp's lab at the University of Nottingham. John is currently focused on human genetics and serves as the ProCardis database manager at the WTCHG in Oxford University.
Key Features of CodonW
Here are some cool features:
- Written in ANSI compliant C
- User-friendly menu-driven interface
- A bunch of command line options available
- Works independently of genetic codes
- No limits on:
- The number of sequences you can analyze
- The length of those sequences
- Calculates various codon usage indices:
- CAI: Codon adaptation index
- Fop: Frequency of optimal codons
- Nc: Effective number of codons
- CBI: Codon Bias Index
- Amino acid indices like:
- GRAVY score:
- Aromaticity:
- An extensive correspondence analysis that includes:
- Correspondence analysis of Codon Usage, RSCU (Relative synonymous codon usage), Amino acid usage
User Reviews for CodonW FOR LINUX 7
-
for CodonW FOR LINUX
CodonW FOR LINUX simplifies Multivariate analysis of codon and amino acid usage efficiently. It offers extensive command-line options for detailed analysis.
-
for CodonW FOR LINUX
CodonW is an outstanding tool for genetic analysis! Its user-friendly interface and powerful features make it a must-have for researchers.
-
for CodonW FOR LINUX
I've been using CodonW for my projects, and it's incredibly efficient! The extensive options available really enhance my analysis capabilities.
-
for CodonW FOR LINUX
CodonW simplifies complex multivariate analyses with ease. I love the detailed reports it generates. Highly recommend!
-
for CodonW FOR LINUX
This app is a game-changer for codon usage studies! The command-line interface offers great flexibility, and the results are impressive.
-
for CodonW FOR LINUX
Absolutely love CodonW! It’s comprehensive yet easy to use, making my research much more manageable. A fantastic resource!
-
for CodonW FOR LINUX
CodonW has transformed how I analyze genetic data. Its robust features and accurate calculations are invaluable to my work!