Description
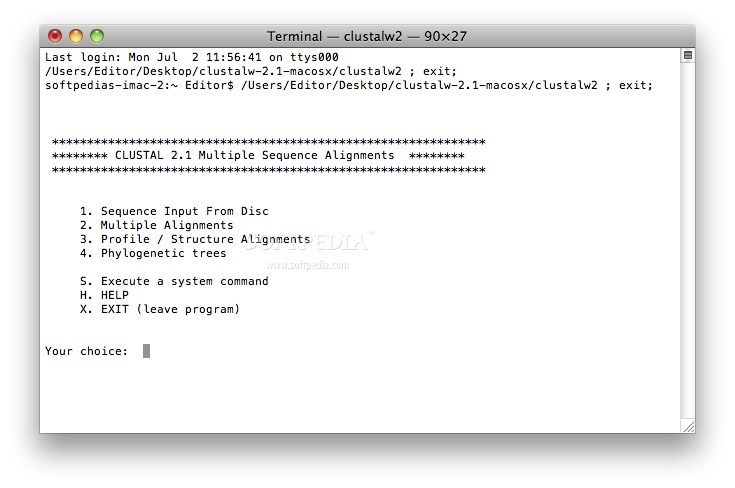
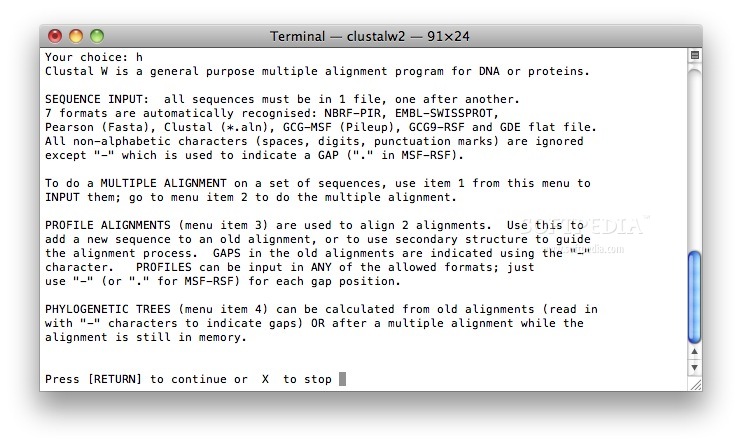
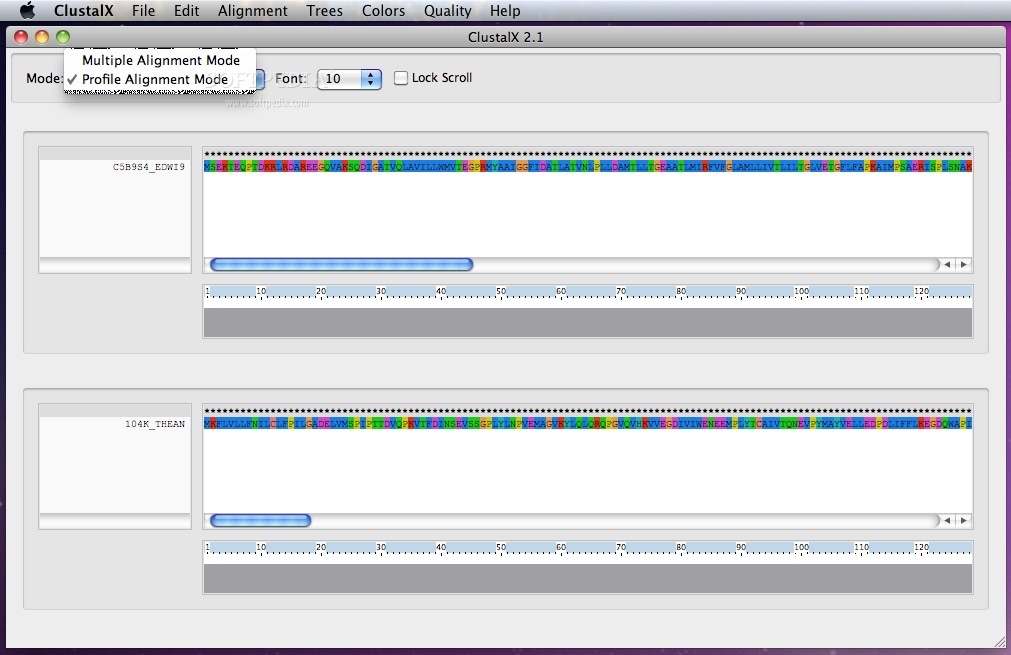
ClustalX is a streamlined OS X utility that provides the necessary tools to align DNA or protein sequences from within a user-friendly interface or a Terminal window.
ClustalX comes with support for numerous input formats, such as GDE, FASTA, NBRF/PIR, GCG9 RSF, Clustal, GCC/MSF and EMBL/Swiss-Prot.
In order to use ClustalX, you have to place each sequence in one file and arrange them one after another.
ClustalX's intuitive interface enables you to perform profile alignments, phylogenetic trees and multiple alignment in just a few easy steps.
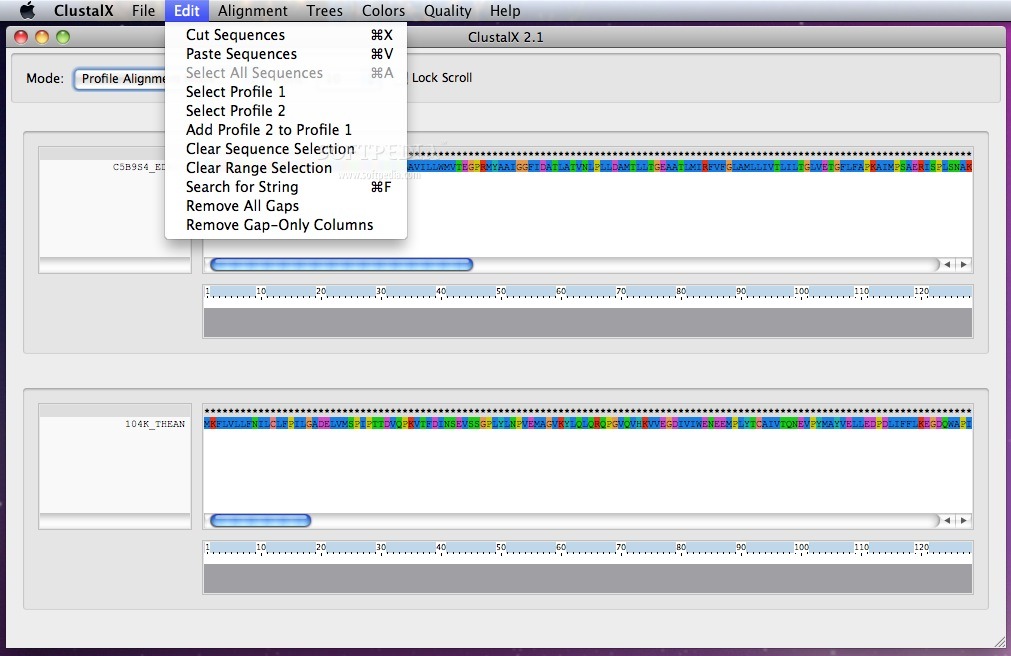
From the Edit menu you can easily search for a String, remove gaps, clear sequences or range selections and switch between profiles.
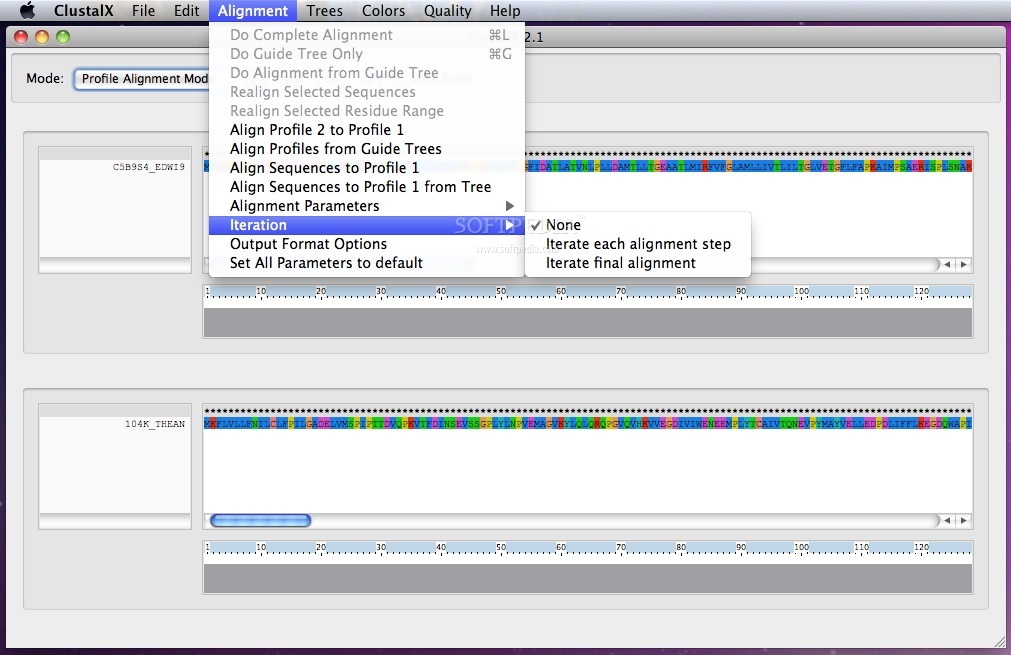
By accessing the Alignment menu, you can easily align the sequences to the desired profile and open the Output Format Option window.
ClustalX also allows you to export to the NEXUS, Clustral, GDE, GCG/MSF and NBRF/PIR format.
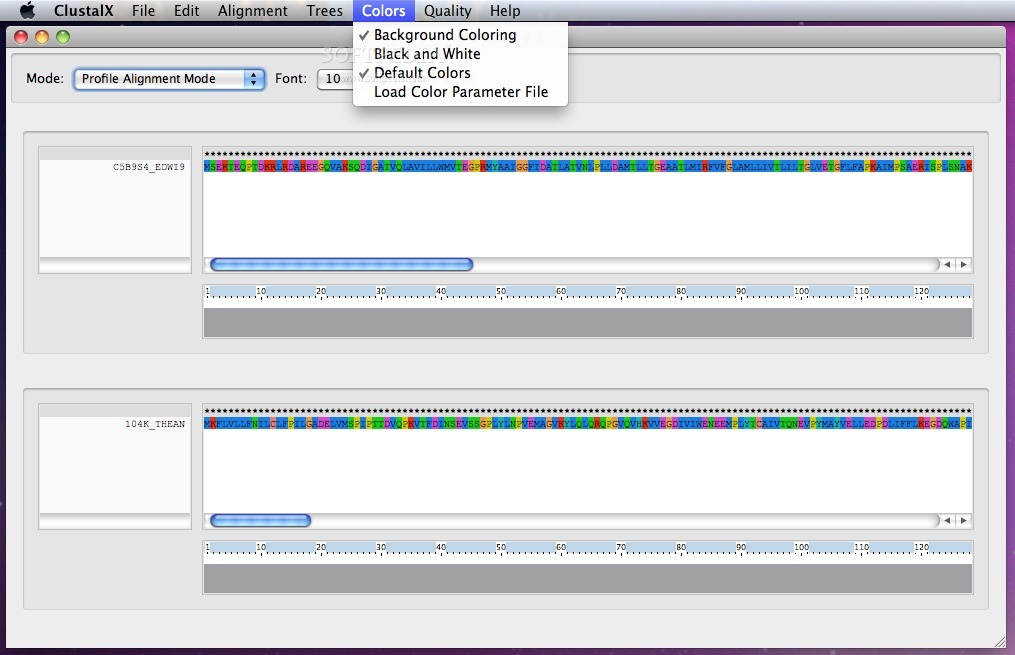
ClustalX enables its users to choose between existing color sets or load a color parameter file with just a mouse click.
On top of that, users who are more comfortable performing all aforementioned tasks from a Terminal window, can download and use ClustalW.
Users that are interested in more advanced features like using secondary structures for profile alignment can use the "Using ClustalX for multiple sequence" tutorial created by by Jarno Tuimala.
User Reviews for ClustalX FOR MAC 1
-
for ClustalX FOR MAC
ClustalX FOR MAC provides a user-friendly interface for aligning DNA or protein sequences, with support for various formats. Great for bioinformatics tasks.