Description
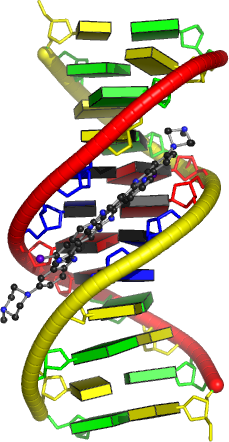
3DNA is a versatile package for analyzing and rebuilding three-dimensional nucleic acid structures, based on a standard reference frame.
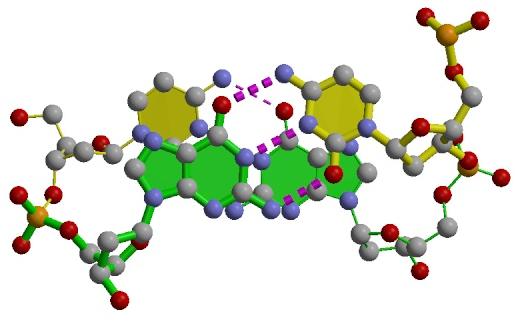
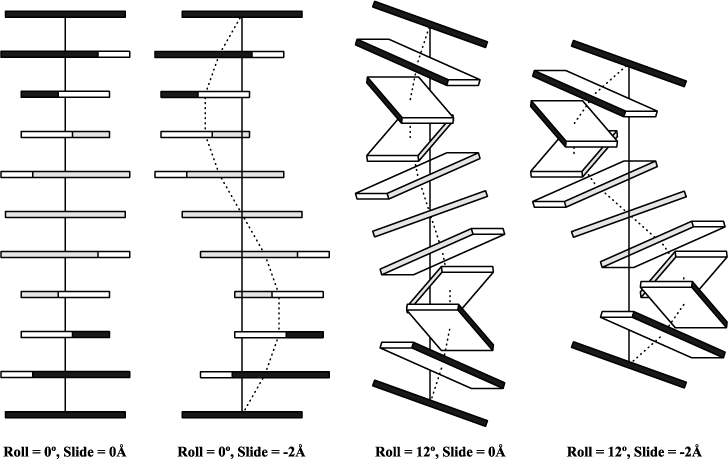
In its core, the software uses a simple, yet mathematically rigorous and geometrically sensible, scheme for calculating a complete set of local base-pair, step, and helical parameters, and allows for exact rebuilding of a structure based on these parameters.
Unique features of 3DNA include automatic classification of a dinucleotide step as A-, B-, or TA-like based on the positioning of the phosphorus atoms, and the generation of "standardized" base stacking diagrams. Calladine-Drew style is given schematic representations as full atomic models with the sugar-phosphate backbone and of DNA as well thanks to the rebuilding routines.
3DNA was created by Dr. Xiang-Jun Lu in the laboratory of Professor Wilma K. Olson at the Department of Chemistry and Chemical Biology, Rutgers-the State University of New Jersey. Since leaving Rutgers, Dr. Lu has continued to work on this project as an independent consultant in his spare time. This updated website provides users with more background information and technical details about 3DNA, including working examples. Thus, those who are really interested in nucleic acid structures can have a clear understanding of "what's going on in that black box".
3DNA has benefited greatly from interactions with the Nucleic Acid Database (NDB) project led by Professor Helen Berman. Dr. Zukang Feng at the PDB/NDB, Dr. A. R. Srinivarsan, Ms. Yurong Xin, Mr. Andrew Colasanti, Mr. Jin Tao, Ms. Fei Xu and other members of the Olson lab., and numerous users from around the globe have helped in making 3DNA a better tool to serve the scientific community.
User Reviews for 3DNA FOR MAC 1
-
for 3DNA FOR MAC
3DNA FOR MAC is a sophisticated tool for analyzing nucleic acid structures. Its unique features and rebuilding routines make it a valuable resource for researchers.