Description
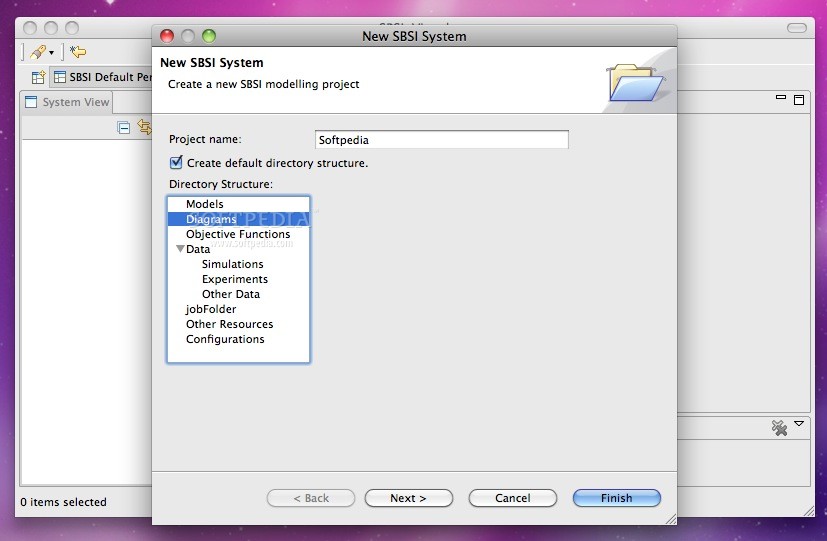
SBSI (Systems Biology Software Infrastructure) is an open source, easy to use suite of tools for systems biology, such as parallelized numerical algorithms, and an Eclipse RCP based client for visualizing results, running simulations, and integrating SBML based plugins.
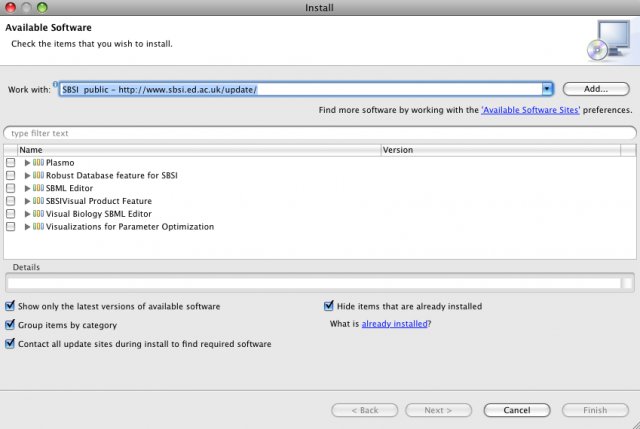
There are three main software components in SBSI: SBSINumerics, SBSIVisual and SBSIDispatcher.
SBSINumerics contains a configurable, parallelized command line application for running parameter estimations.
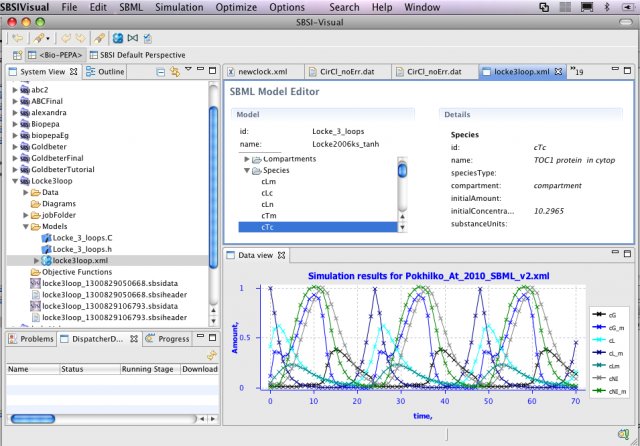
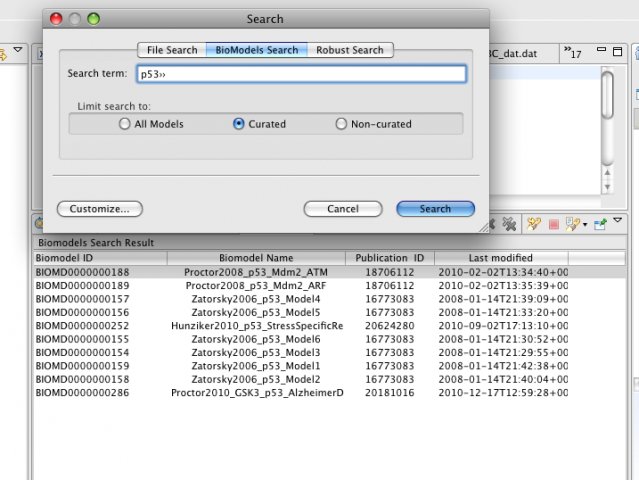
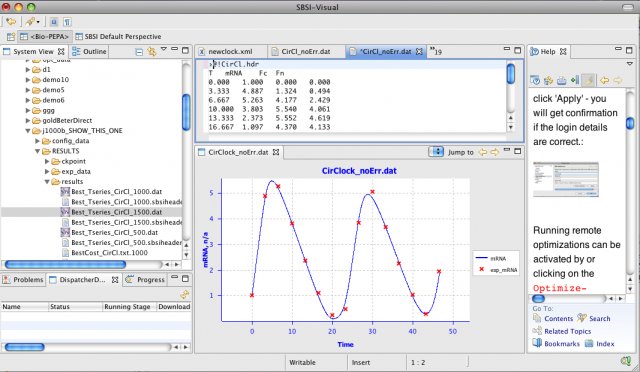
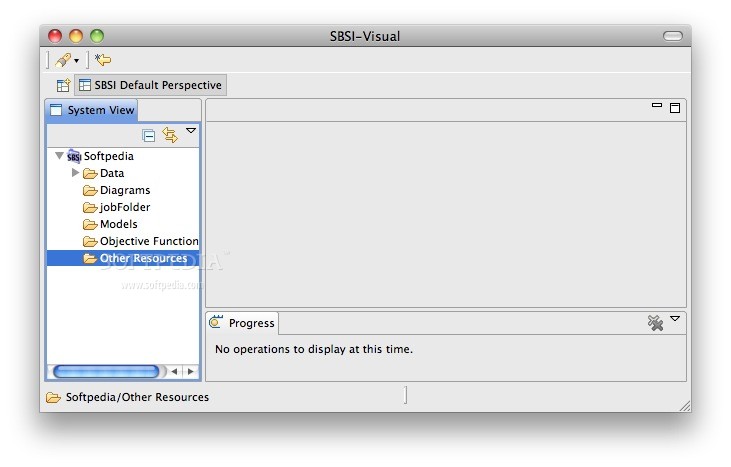
SBSIVisual is a desktop application, that provides easier access to SBSINumerics functionality for non-programmers, and can display results, run simulations, and interact with systems biology databases.
SBSIDispatcher is a middleware component, that can deploy and manage SBSINumerics procedures on backend servers.
User Reviews for SBSI FOR MAC 1
-
for SBSI FOR MAC
SBSI FOR MAC is a robust suite for systems biology. SBSIVisual is user-friendly for non-programmers, and SBSIDispatcher efficiently manages procedures.