Description
Progenesis Post-Processor was designed to allow the results from Progenesis to be converted into mzQuantML for label-free methodologies, and subsequently extended to cover label-based and top3 quantitation.
In other words, Progenesis Post-Processor is a Java based tool to enable the feature-level quantitation performed by Progenesis to be used for some other techniques, such as SILAC.
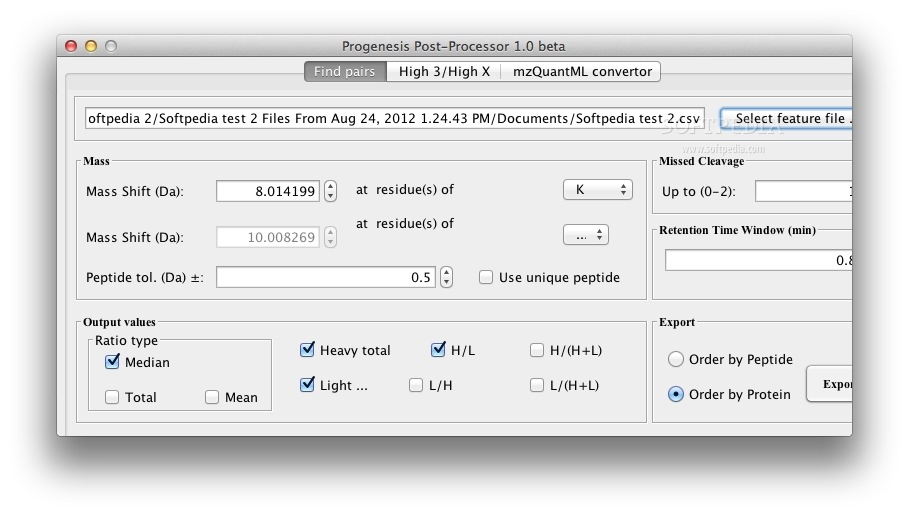
Progenesis Post-Processor searches the table of features to find pairs of features nearby in the RT dimension of the 2D map, separated by the mass shift in the m/z space due to the heavy label being incorporated into lysine or arginine.
For instance, typical labels incorporate 13C and/or 15N into lysine and arginine to produce known mass shifts of +6Da, +8Da, +10Da, and so on.
There are no built-in limits on the mass shift that is permissible, for example overlapping features (say +2Da) are in theory identifiable, so long as Progenesis LC-MS has correctly separated features in the 2D map, although this feature has not yet been comprehensively tested.
Progenesis Post-Processor is a cross-platform utility capable of running on any operating system that comes with Java support (e.g. Mac OS X, Windows, Linux).
User Reviews for Progenesis Post-Processor FOR MAC 1
-
for Progenesis Post-Processor FOR MAC
Progenesis Post-Processor is a powerful Java-based tool that enables feature-level quantitation for various techniques like SILAC. Great for converting Progenesis results to mzQuantML.