Description
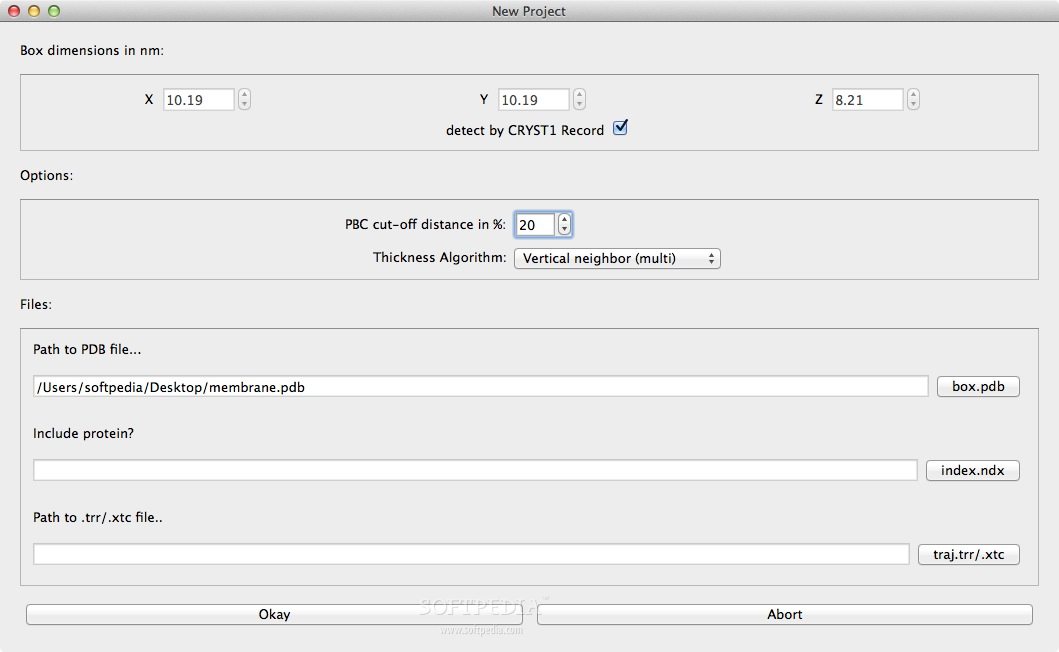
APL@Voro is a straightforward OS X application specially designed to analyze GROMACS Molecular dynamics trajectories of lipid bilayer simulations.
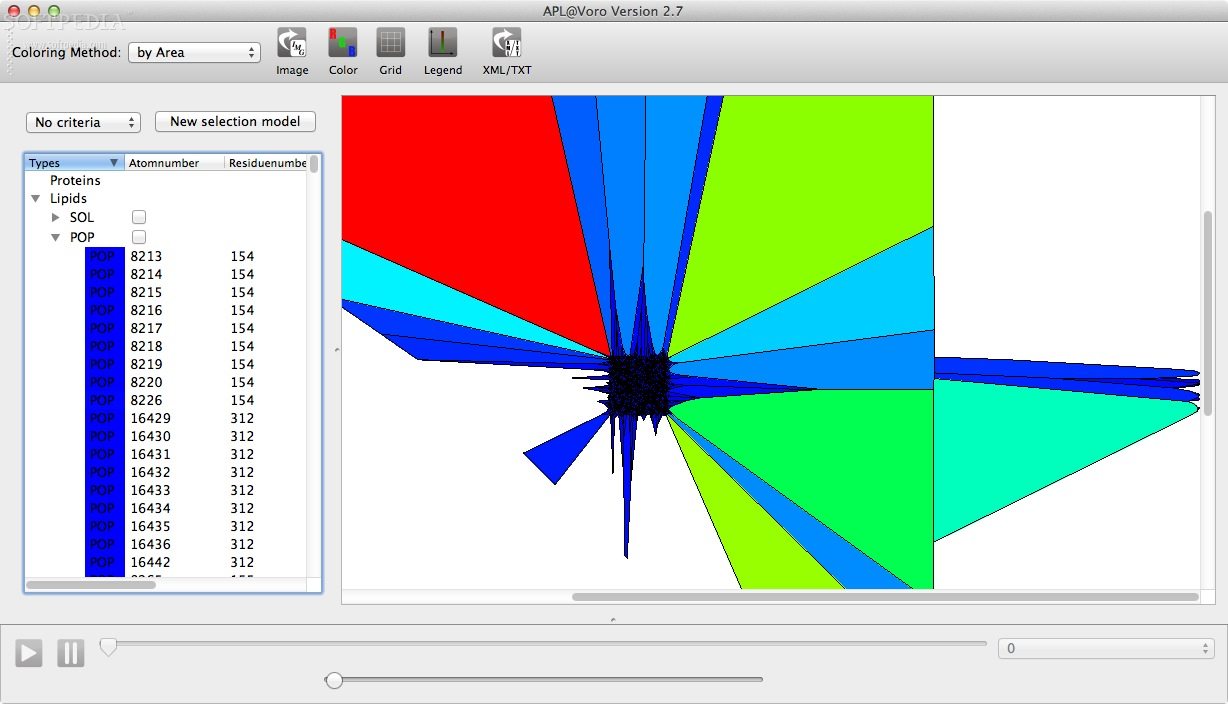
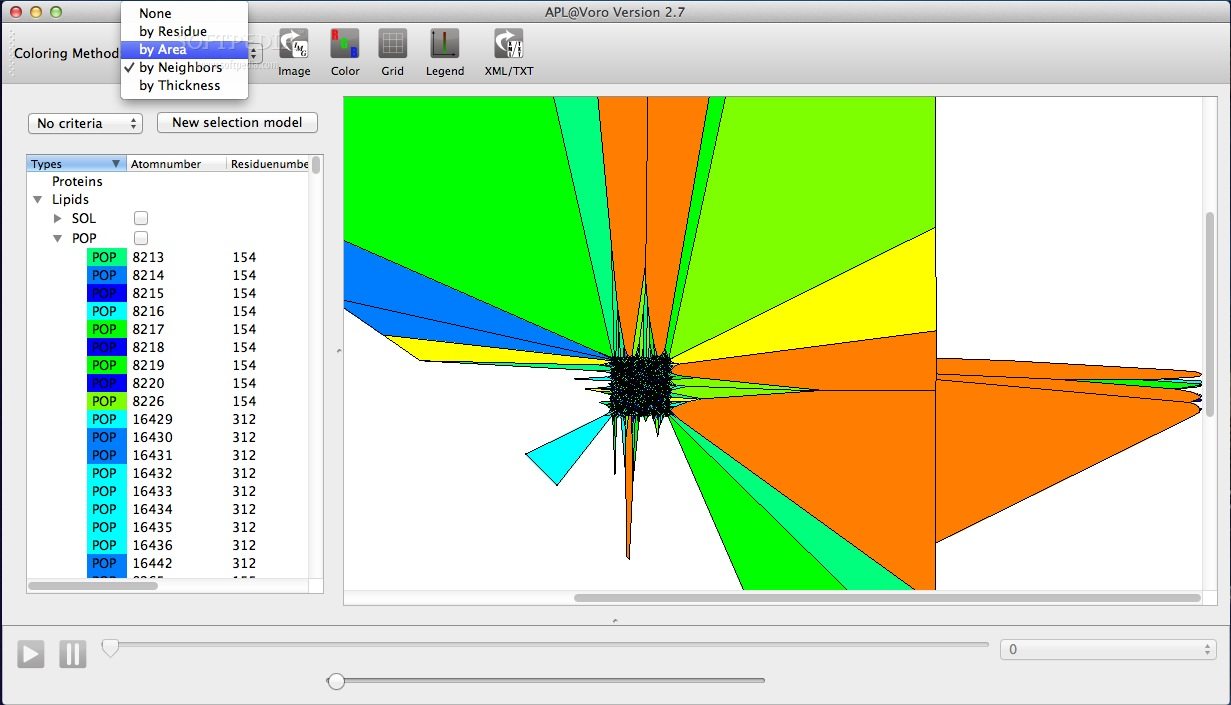
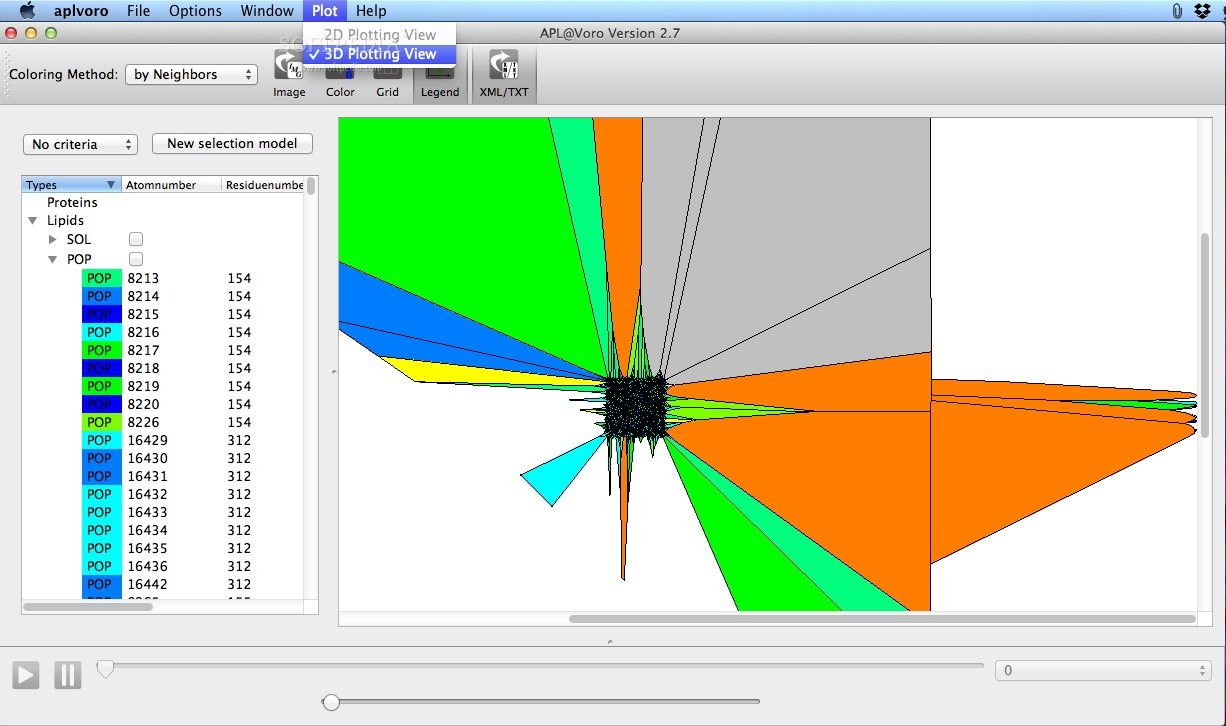
APL@Voro is able to load and analyze PDB coordinate files, GROMACS trajectory files, and a GROMACS index files to create a two dimensional geometric representation of a bilayer.
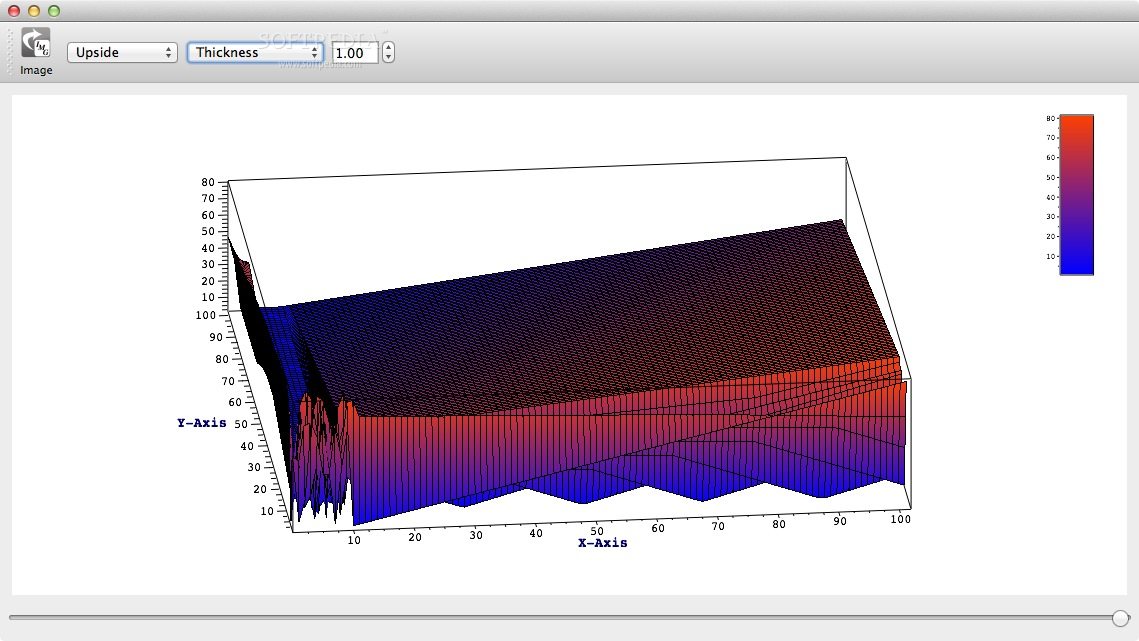
APL@Voro comes with support for complex bilayers with a mix of various lipids and proteins and is capable to calculate the area per lipid and the bilayer thickness.
User Reviews for APL@Voro FOR MAC 1
-
for APL@Voro FOR MAC
APL@Voro FOR MAC provides an intuitive interface for analyzing lipid bilayer simulations. Impressive support for complex bilayers, lipid calculations and geometrical representation.